library(tidyverse)
library(magrittr)
library(plotly)
data <- china_annual_data %>% pivot_longer(cols=2:5, names_to = c("type"), values_to = c("count"))
data
# A tibble: 64 × 3
year type count
<int> <chr> <dbl>
1 2005 dengue.cases.indigenous 0
2 2005 dengue.cases.imported 45
3 2005 counties.with.dengue.fever.indigenous 0
4 2005 counties.with.dengue.fever.imported 33
5 2006 dengue.cases.indigenous 1007
6 2006 dengue.cases.imported 46
7 2006 counties.with.dengue.fever.indigenous 15
8 2006 counties.with.dengue.fever.imported 33
9 2007 dengue.cases.indigenous 481
10 2007 dengue.cases.imported 56
# ℹ 54 more rows
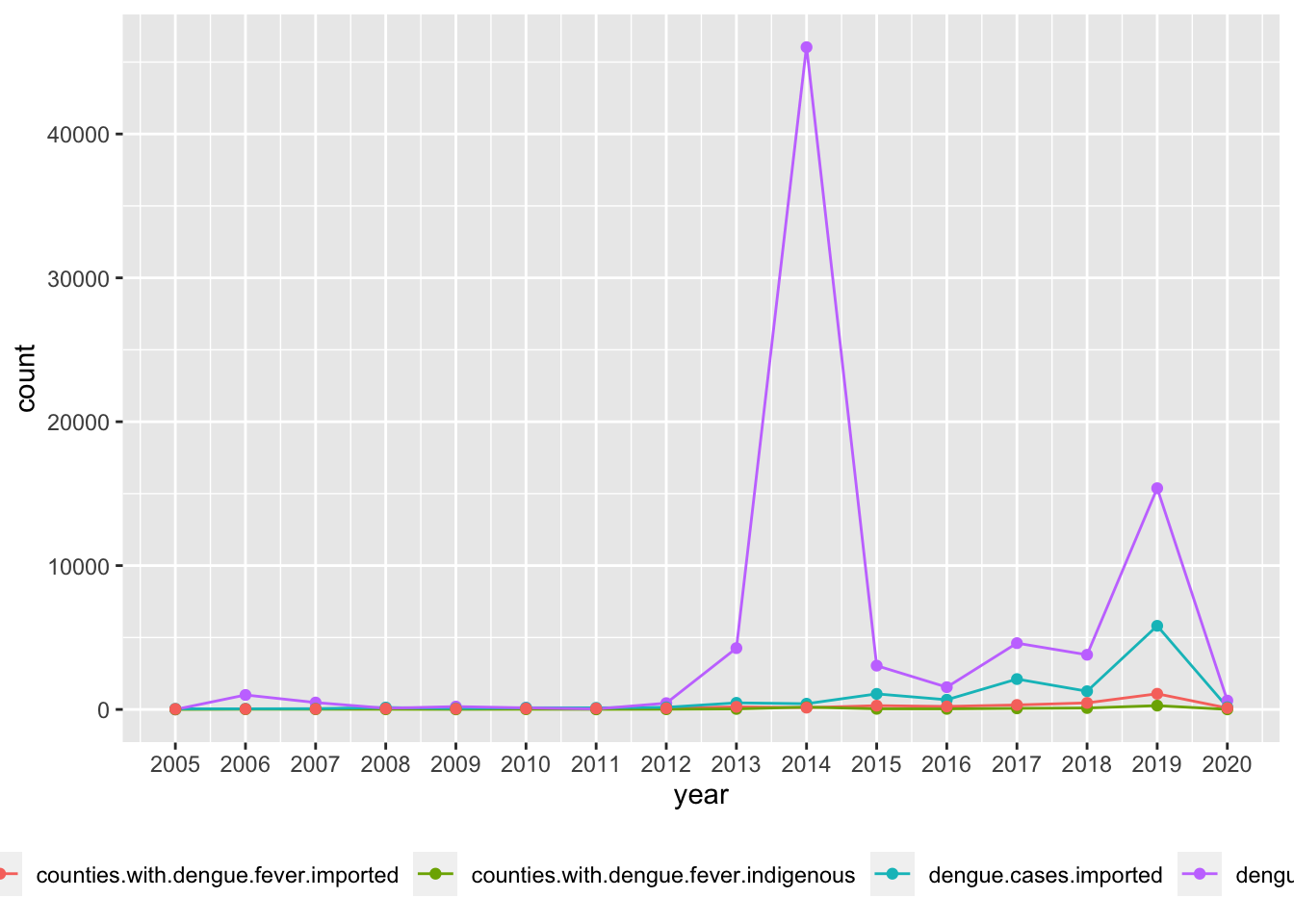
p1 <- ggplot(data, aes(x=year, y=count, group=type)) +
geom_line(aes(col=type)) + geom_point(aes(col=type)) +
scale_x_continuous(breaks=seq(2005, 2020, 1)) + scale_fill_brewer(palette = "Dark2") + theme(legend.position = "bottom")
#plotly::ggplotly(p1) %>%
# layout(legend = list(
# orientation = "h"
# )
# )
p1